Overview¶
Tibanna is a software tool that helps you run genomic pipelines on the Amazon (AWS) cloud. It is also used by 4DN-DCIC (4D Nucleome Data Coordination and Integration Center) to process data.
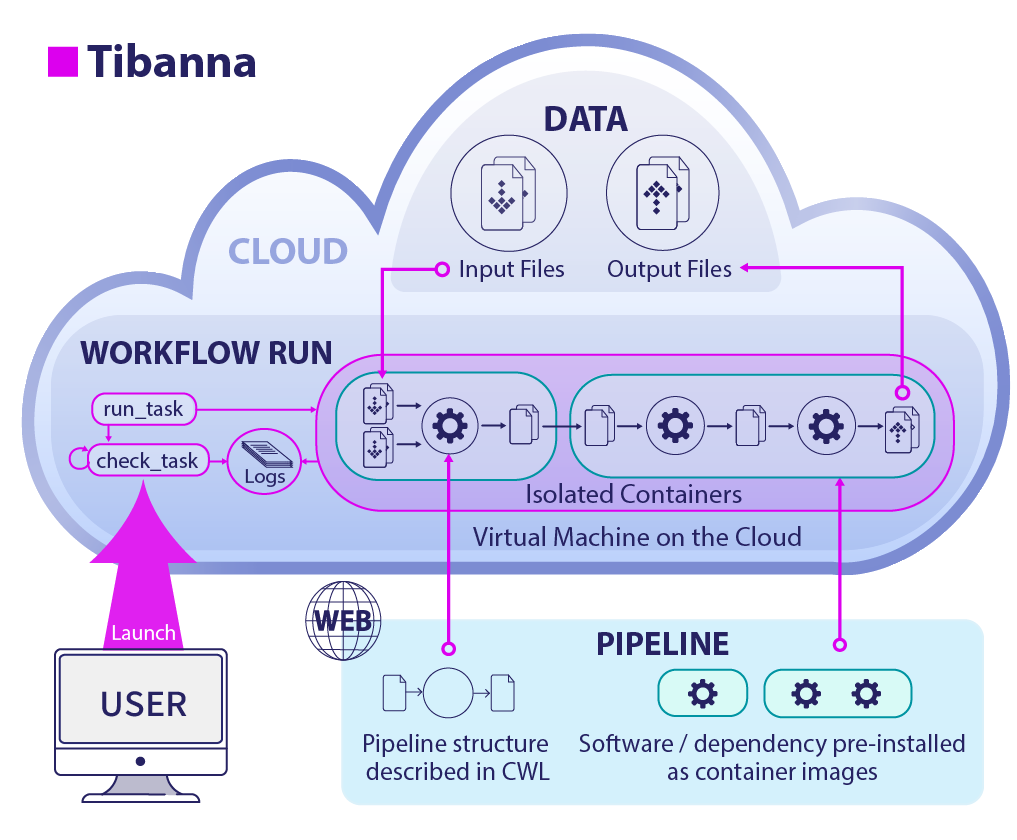
What do I need to run pipelines using Tibanna?¶
- Your pipeline
- Your data
- An Amazon Web Services (AWS) cloud account
- Tibanna
Pipeline¶
- The commands to run your pipeline must be written in either Common Workflow Language (CWL) (recommended), Workflow Description Language (WDL) (only basic support), Snakemake or a list of shell commands.
- The pipelines may be local CWL/WDL/Snakemake files or they should be downloadable from public urls.
- With the exception of Snakemake, your pipeline and dependencies must be pre-installed as a Docker image (https://www.docker.com/). For Snakemake, conda can be used instead.
Data¶
- Your data must be in AWS S3 buckets.
Tibanna¶
- Tibanna is open-source and can be found on github.
- Once installed, Tibanna can be run either as a set of commend-line tools or a set of python modules.
Command-line tools
$ tibanna run_workflow --input-json=run1.json
Python
>>> from tibanna.core import API
>>> API().run_workflow(input_json='run1.json') # input_json is a json file or a dict object
Contents:
- News and updates
- Simple Example Pipeline
- Check Before using Tibanna
- Installation
- Command-line tools
- Python API
- Job Description JSON Schema
- Monitoring a workflow run
- Common Workflow Language (CWL)
- Workflow Description Language (WDL)
- Snakemake
- Amazon Machine Image
- Running 4DN pipelines using Tibanna
- How it works